CUSABIO는 암, 세포 생물학, 면역학, 신경과학, 후생유전학 등 연구 분야의 글로벌 고객에게 60,000개 이상의 검증된 항체, 10,000개 이상의 재조합 단백질, 660개 이상의 사이토카인 및 수천 개의 ELISA 키트를 제공하는 검증된 제조업체 입니다. 다양한 과학 연구 분야에서 고객의 폭 넓은 요구를 충족시킬 수 있는 Cusabio의 제품을 만나보세요.
Western blotting uses antibodies to identify individual proteins from complex samples and to perform a semi-quantitative analysis. First, proteins are separated from each other based on their size by SDS-PAGE. Next, the proteins are transferred from the gel to membrane by application of an electrical current. Finally, the antigen-loaded blotting membrane could be detected and analyzed according antigen-antibody specific binding by a specific primary antibody.
Western blotting is mainly used for qualitative or semi-quantitative analysis of target protein-specific expression, subsequent analysis of protein-protein or protein-DNA interaction, and identification analysis of protein modification.
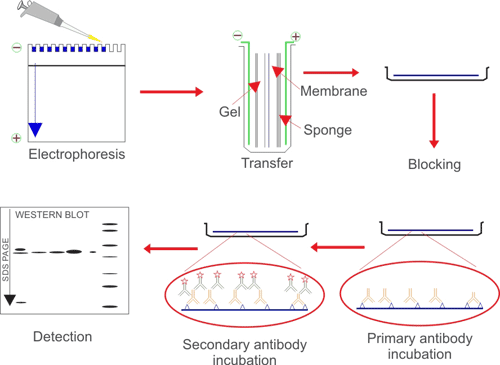
Fig. 1. The brief processes of WB
1. Protein Sample Preparation
1.1 Source of Protein Sample
Protein samples for western blotting can be soluble protein fluids, cell/tissue lysates or immunoprecipitated proteins. The protein loading differs from different samples, basically, the recommended protein loading of purified protein is no more than 100 ng, and the loading of cell/tissue lysate could be 10-40 μg.
1.2 Protein Sample Preparation
Generally, complex protein components are extracted from animal or plant tissues or cells, and the following principles should be observed during the extraction process:
a. Decide the appropriate extraction method based on the characters of individual protein.
b. Use the appropriate method to maximize the extraction of target protein.
c. Perform under low temperature and add protease inhibitors to prevent protein degradation.
d. Choose the appropriate protein lysate to maintain protein solubility.
Store Protein samples at -80℃, avoid repeated freezing and thawing, detect as soon as possible.
Preparation of lysate from cell culture
a. After the cell confluence reaches 80%, place the cell culture dish on ice and wash the cells with ice-cold PBS for 3 times.
b. Prepare lysates which containing protease inhibitors. The commonly used protease inhibitors are shown in the table below (Table 2). Appropriate protease inhibitors should be selected according to the experimental requirements. The most commonly used protease inhibitor is PMSF (working concentration is 1 mM), which is highly toxic, so it should be self-protected when used. Its half-life in water is extremely short, so it should be added before use.
c. Add 1 mL of protein lysate containing protease inhibitor to a 10 cm culture dish, shake gently, and lyse on ice for 15-30 min.
d. Scrape adherent cells off the dish using a cold plastic cell scraper, then gently transfer the cell suspension into a 1.5 mL EP tube, then place the tube on ice. Bubbles should be avoided at this time.
e. Centrifuge at 12000 rpm for 10-15 min at 4℃.
f. Gently aspirate the supernatant to another fresh tube and place on ice for later use. Be careful not to absorb impurities such as lipids floating in the upper layer.
g. After protein quantification, add appropriate amount of 6 × sample loading buffer, and boil at 95℃ for 5 min, then centrifuge at 12000 rpm for 30 sec, lastly, store at -20℃.
Sample preparation notes:
All steps must be operated at low temperature! Low temperature! Low temperature!
a. For the cells grown in suspension, collect by centrifugation at 2500 rpm for 3 min, followed by cell washing and lysis procedures.
b. For drug-treated cells, especially samples from apoptosis related studies, media supernatants should also be collected.
c. It is not recommended to use protease to digest and collect cells. Because it may introduce protein impurities or cause damage to some certain proteins, especially the membrane surface proteins, to interfere the experimental results.
d. A viscous transparent gel may appear in the lysate. The transparent gel is a genomic DNA component. Take the supernatant for experiments. However, when the target protein is tightly bound to the genome, the gel needs to be ultrasonically disrupted or syringe-sucked, then take supernatant for subsequent experiments to avoid protein loss.
e. PMSF is unstable in aqueous solution, usually it degrades by half in 30 min. The rate of loss of activity increases with the increase of pH value, and the deactivation rate at 25℃ is higher than 4℃. When the sample is processed for more than 1 h, it needs to be added once more.
f. Pay attention to the influence of cell state and the number of cell passages. Heterogeneity exists in cancer cells of different algebras, so the cell morphology, migration and invasion ability may change, thereby make some gene expression change as well.
On one hand, due to the certain heterogeneity of the cells themselves, after a period of cultivation, the overall characteristics of the cells are gradually changed in a way of survival of the fittest.
On the other hand, in the process of cell culture, due to changes in culture conditions or the presence of external stimuli, such as replacement of culture reagents, digestion and passage, cell contamination, and some chemical and physical stimuli, the expression of related genes in cells may be affected. Ultimately affect the experimental results.
When using tumor cells for experiments, it should be preserved first, and try to use relevant cells in the same algebra to carry out relevant experimental research to avoid the occurrence of cell heterogeneity due to excessive passage times, and ultimately lead to inconsistencies in experimental results.
Preparation of lysate from tissues
a. Collect fresh samples and wash with saline or PBS, then cut into appropriate sizes. You can use a 1-2 mL homogenizer for tissue homogenate on ice, or add liquid nitrogen for grinding. It is recommended to use liquid nitrogen grinding, for the tissue block is not easily damaged and there is frictional heat generation during the homogenization process.
b. Prepare the lysate containing protease inhibitor.
c. Add appropriate amount of lysate containing protease inhibitor (50 mg/500 μL) into the grinded tissue sample, and place the tube on ice for 15-30 min for lyse, meanwhile, intermittently mix to fully lyse.
d. Centrifuge at 12000 rpm for 10-15 min at 4℃.
e. Remove the EP tube gently, and absorb the supernatant into a fresh tube. Be careful not to absorb impurities such as lipids floating in the upper layer, then place on ice for later use.
f. Centrifuge at 12000 rpm for 10-15 min at 4℃.
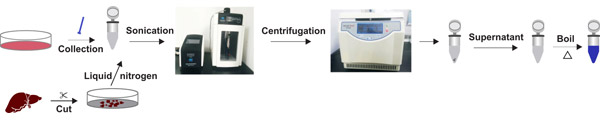
Fig. 2. The processes of lysate preparation
1.3 Selection of Protein Lysate
For most samples, RIPA lysis buffer can be used for rapid cell lysis.
Table 1. The components of RIPA lysis buffer
| RIPA Lysis Buffer | |
|---|---|
| Tris-HCl | 50 mM |
| NaCl | 150 mM |
| EDTA | 1 mM |
| SDS (W/V) | 0.1% (W/V) |
| Sodium deoxycholate | 1% (W/V) |
| Triton X-100 | 1% (V/V) |
| Appropriate protease inhibitors can be added to the RIPA lysis buffer according to the purpose of experiment. | |
The main components of the protein lysate and their effects are as follows:
Buffer
A buffer with a certain pH range could provide a stable environment for proteins and increase protein solubility as well. The buffers of Tris-HCl or HEPES, pH 7.4 with similar physiological pH are commonly used. The buffer of Tris-HCl (pKa = 8.1) has a pH range of 7.0-9.2, which is sensitive to temperature. The pH value range of HEPES (pKa = 7.55) is 6.5-8.5.
Saltion